Tadahaya MIZUNO
Assistant Professor,
University of Tokyo and Institute of Statistical Mathematics, Japan
Adjunct Lecturer, Department of Bioregulation, Graduate School of Medicine, Nippon Medical School
About Us
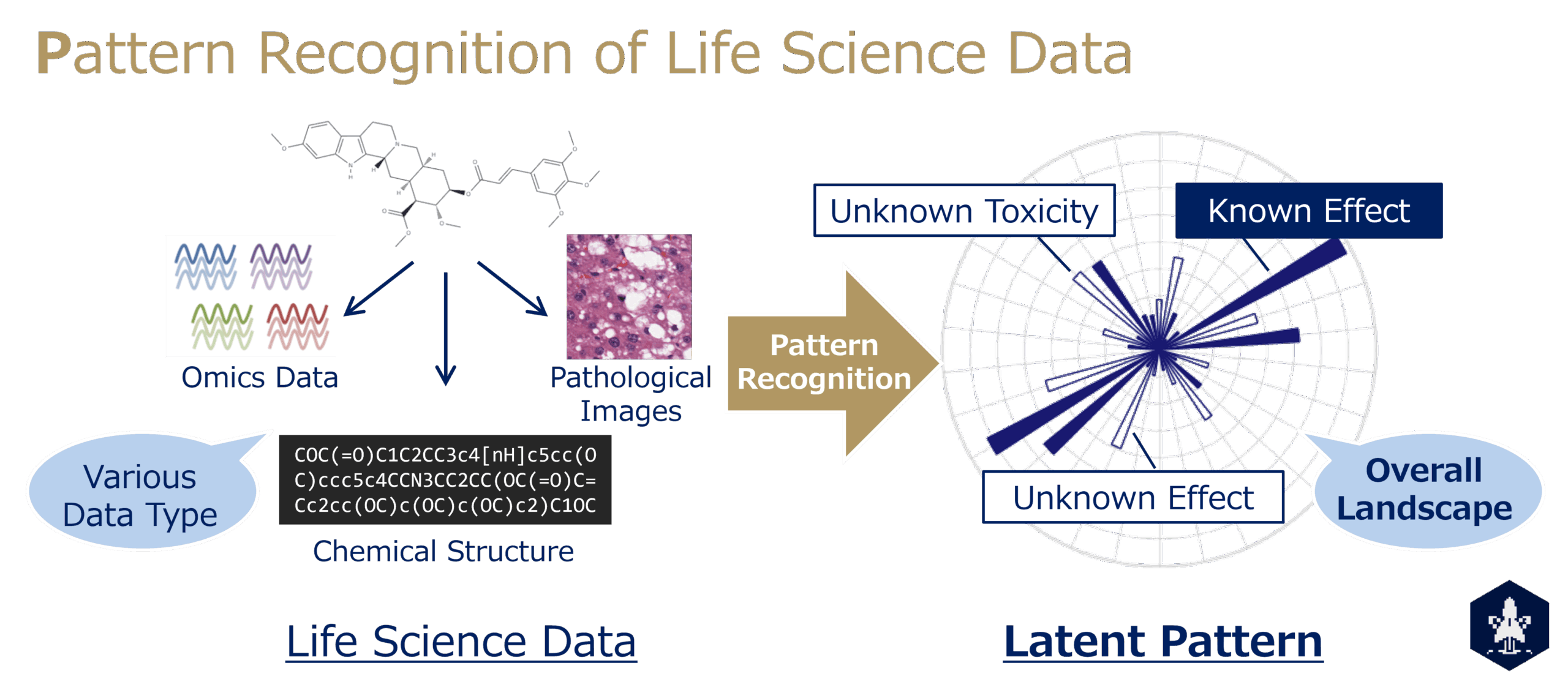
How can we recognize structures that cannot be directly observed?
Our team aims to expand the framework of understanding in life sciences by uncovering patterns and structures hidden behind complex data through mathematical modeling. Focusing on pharmaceutical science as a central theme, we develop and apply deep learning and statistical methods to extract and visualize unbiased representations, bringing to light essential structures embedded in observational data.
News
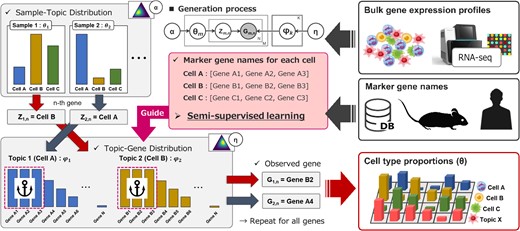
GLDADec: marker-gene guided LDA modeling for bulk gene expression deconvolution
Inferring cell type proportions from bulk transcriptome data is crucial in immunology and oncology. Here, we introduce guided LDA deconvolution (GLDADec), a bulk deconvolution method that guides topics using cell type-specific marker gene names to estimate topic distributions for each sample.
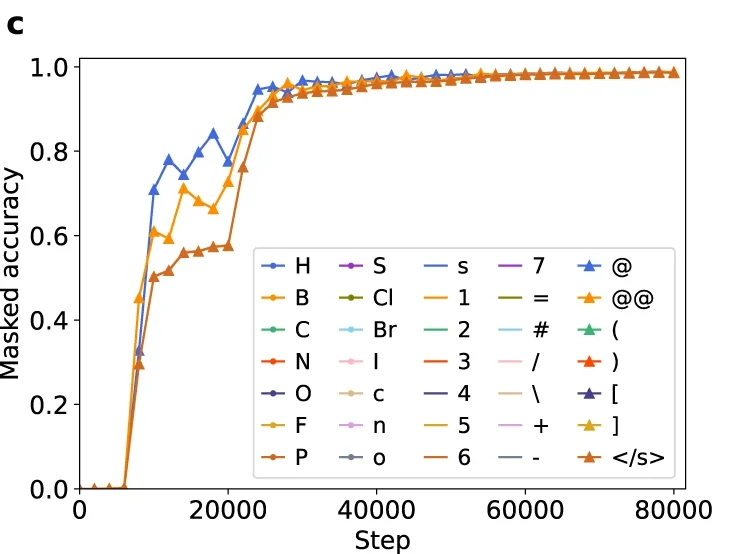
Difficulty in chirality recognition for Transformer architectures learning chemical structures from string representations
We found that the Transformer requires particularly long training to learn chirality and sometimes stagnates with low performance due to misunderstanding of enantiomers.
Projects
Our main research projects
Environments
Our research environments