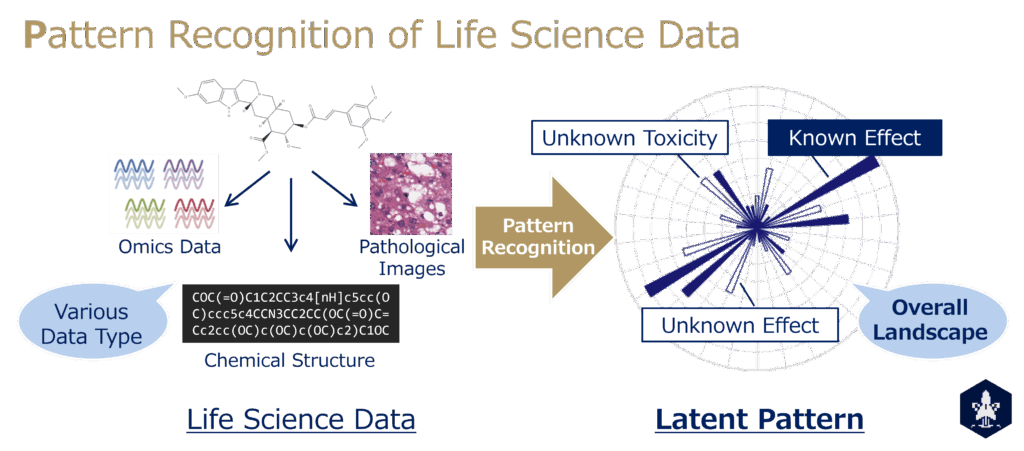
Research
Unexpected adverse events leading to market withdrawal and the success of drug repositioning reveal that medicines often harbor unknown aspects not anticipated even by their developers. Our research aims to systematically capture these “unrecognized dimensions” through pattern recognition of life science data. Specifically, we seek to extract latent and essential patterns from diverse data types—including omics data, chemical structures, toxicopathological images, and scientific literature—and to represent and visualize them mathematically, thereby enabling a comprehensive understanding of drug properties.
A central requirement in this effort is the minimization of arbitrariness in data acquisition. We view this from two perspectives. The first is comprehensive data, exemplified by omics analyses, where variables belonging to a particular layer are collected exhaustively, allowing unbiased quantification within that layer. The second is sensory-proximal data, exemplified by images, which can be acquired with minimal human intervention and thus provide information with reduced subjectivity.
Such data often contain redundancy and invariance, making it difficult to directly extract meaningful features. Here, pattern recognition becomes essential. By employing a range of methods—including latent representation learning with deep learning and statistical frameworks such as latent variable models—we aim to capture the underlying structures of data in a holistic manner. This is the core subject of our research. We explicitly pursue the question: “For a given input (data), what methods yield what outputs (patterns)?”
We first demonstrated the validity of this strategy in vitro through transcriptome analysis of compound-treated cells (Nemoto S., J Nat Prod., 2021), and we are now extending it to in vivo and in human studies. Recent achievements include the application of these approaches to omics, toxicopathological images, chemical structures, and text data, advancing toward the establishment of methodologies for uncovering the hidden aspects of drugs (Morita K., Toxicol Sci., 2023; Maedera S., Comput Biol Med., 2024; Yoshikai Y., Nat Commun., 2024). Collectively, this line of research seeks to expand human cognition and provide a universal framework that facilitates the social integration of scientific knowledge.
Education
In addition to research, we place strong emphasis on cultivating the next generation of researchers. A key advantage of approaching data science from a pharmaceutical background is the ability to understand both wet experiments and in silico analysis. Pharmaceutical sciences inherently foster interdisciplinarity, as diverse experts collaborate toward the shared goal of drug discovery.
What matters here is not becoming someone who merely “handles data,” but rather developing as a researcher who can generate discoveries by bridging pharmaceutical sciences and data science. We are committed to training such individuals who can integrate the two fields to open new perspectives in both basic and applied sciences.
If you are interested in taking on bioinformatics, applying pharmaceutical knowledge in new ways, or connecting data science to experimental research, please feel free to contact us. We provide an environment not only for scientific training but also for thinking together about your future career.